The Preprocessing Tool lets you specify a unique pre-processing protocol and execute it in one step. There are many preprocessing operations included in the Preprocessing tool that may be added to the protocol.
Getting There
The Preprocessing Tool is available via the Tools Menu.
Function
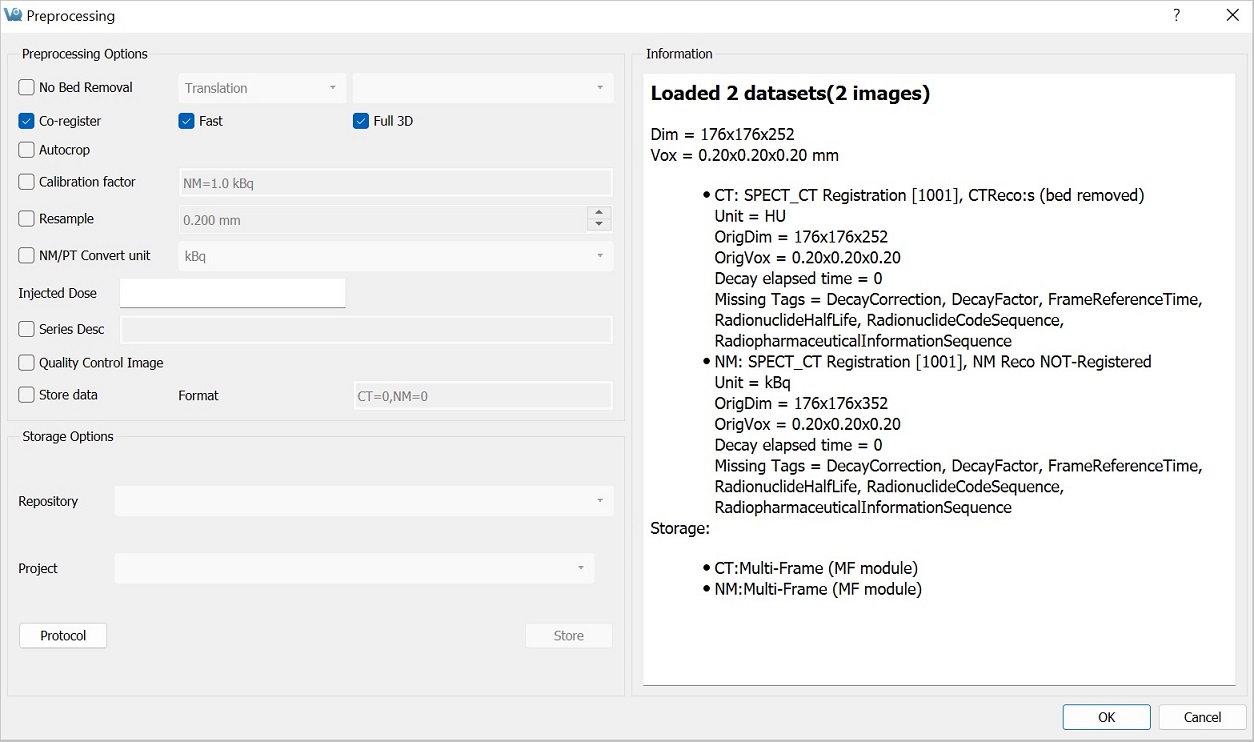
The Preprocessing Tool panel is divided into three sections. The first section is the Preprocessing Options section, which includes the preprocessing operations that may be included in the protocol.
| CT Bed Removal | Automatically detects the bed position within the image and removes the CT voxels which constitute the animal bed. (Only supports Minerve beds). |
| Co-register | Performs automatic image co-registration of all inputs to the Reference. If Fast or Full 3D are not checked, then Standard and Translation are used respectively. |
| Autocrop | Performs automatic cropping. Search for boundaries starts at the end slices of the image and moves towards the center of the image until a non-background voxel is detected from all 6 faces. |
| Calibration Factor | Applies a scalar multiplication to NM data (PET/SPECT). It is important to pay attention to the formatting of the input cell. Calibration factors per modality [comma separated] in format - “Mod=1.0 Unit”. Example: “PT=13456.7 BQML”. The case of the unit should match what is listed in the Information window. |
| Resample | Up/Downsamples voxels to the specified pixel dimensions. |
| NM/PT Convert unit | Converts supported units to/from uptake and/or concentration units. If converting to/from SUV, please provide the Injected Dose information. You miust specify units. Default is [kBq]. |
| Series Description | Appends text to end of series description for all datasets. |
| Quality Control Image | Generates and stores quality control image of preprocessed image. Results stored in the project webdisk of the repository specified below. |
| Store Data | Saves image data being preprocessed to specified iPACS repository below. |
| Format | Specifies formats in which the data will be saved. The numbers 0, 1, and 2 correspond to multi-frame (MF module), multi-frame (MF function groups), and single frames (one Z slice per file) formats, respectively. For multiple data sets, formats are separated by a comma. The current formats of the loaded data sets will be displayed in this section upon opening the tool as well as in the Information section. |
The second section is Storage Options, where users may designate the iPACS repository and Project folder where preprocessed datasets will be saved (note: Store data must be checked).
| Repository | Specifies the repository. Repositories can be configured in the DICOM tab of the Configuration window. See the DICOM settings page for more information. |
| Project | Specifies the sub-project (iPACS) or sub-folder (local path) to which data will be saved. |
| Protocol | Copies and/or pastes preprocessing protocol settings to be used at a later time. Modifications can be made directly within the Protocol window. |
| Store | Store Image data to the specified location. |
The final section is the Information section, which features information about the current state of datasets loaded in VivoQuant. Included in this information is the image dimensions and voxel size for the datasets as a whole, as well as basic header metadata about each dataset specifically.
To execute the pre-processing protocol, click OK.